Introduction
In this post I’ll talk about my largest project in terms of time. It started in the middle of the year 2000 as my Master thesis subject and ended up extending itself until around mid 2013 (although it remained in hibernation for almost 10 years in between). The project was an “automatic cell counter”. Roughly speaking, it is a computer program that takes an image with two kinds of cells, and tells how many of each kind of cell there is in the image. There are some details about the “cells” because technically we count nucleus. Also, they are the same “kind” of cell except that some have the presence of a protein an others don’t, etc… In this post we will abstract out the pathological details and pretend we are counting cells of different kinds.
At the time, my advisors were professor Adrião Duarte and Agostinho Brito (two of the nicest people that exists is on this planet). Also, I had no idea about the biology side of things, so I had a lot of help from Dr. Alexandre Sales and Dra. Sarah Jane both from the “Laboratório Médico de Patologia” back in Brazil. They were the best in explaining to me all the medical stuff. Also, they provided a lot of images and testing for the program.
The “journey” was an adventure. From publishing my first international paper, going to Hawaii on my first plane flight, down to asking for funding to buy a microscope in the local financing agency (FAPERN). Although this won’t be a fully technical post, the goal here is to talk about the project itself and things related to it during the development phase.
The problem
The motivation for the project was the need for cell counting in biopsies exams. The process goes more or less like this; When you need a biopsy of some tissue, the pathologist prepares one or more laminas with very very thin sheets of the tissue. The tissue in those laminas are coloured with a special pigment. That pigment has a certain protein designed to attach itself to specifics substances present in the cells that are behaving differently like ongoing division, for instance. The result is that the pigment makes the cells (actually the nucleus) to appear in a different colour depending on the pigment used. Then, we have means to differentiate between the two “kinds” of cells in the lamina.
The next step is to count the cells and compute how many of them is coloured and how many are not. This ratio (percentage of coloured vs non-coluored) will be the result of the biopsy. The problem here is exactly the “counting” procedure. How the heck does one count individual cells? The straight forward way to do that is to put the lamina into the microscope, look into the objective and, patiently and with caution, start counting by eye. Here we see a typical image used in this process.
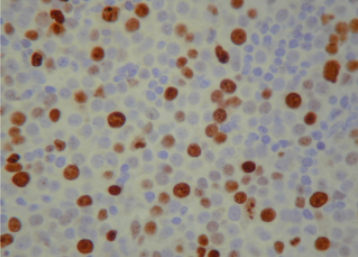
There are hundreds of nucleus (cells) in those kinds images. Even if you capture the image and count them in a screen, the process is tedious! Moreover, the result can’t be based on one single image. Each lamina can produce dozens or hundreds of images that are called fields. To give the result of an exam, several fields have to be counted.
Nowadays, there are machines that do this counting but they are still expensive. At the time I did the project, they were even more expensive and primitive. So, any contribution to this area that would help the pathologist to do this counting more precisely and easily would be very good.
The App
The solution proposed in the project was to make an application (a computer program at the time) that would do this counting automatically. Even at the time, it was relatively easy to put a camera into a microscope. Today it is almost a default 😄. So, the idea was to capture the image from the camera coupled in the microscope, give it as input to the program, and the program would count how many cells are in the image.
Saying it like this, makes it sound simple… But it is not trivial. The cells, as you can see in the previous sample image, are not uniformly coloured, there is a lot of artefacts in the image, the cells are not perfect circles, etc. Hence, I had to device an algorithm to perform this counting.
The algorithm is basically divided in 3 main steps that are very common in image processing solutions;
1 – Pre-Processing
2 – Segmentation
3 – Post-Processing
4 – Counting
Algorithm
The pre-processing stage consisted in applying a colour transform and performing an auto-contrast procedure. The goal was to normalise the images for luminance, contrast, etc. The segmentation step consisted in label each pixel as one of three categories: cell 1, cell 2 or intra-cellar medium (non-cell). The result would be an image where each pixel is either 1, 2 or 3 (or any arbitrary triple of labels). After that comes the most difficult part, which was the post-processing. The segmentation left several cells touching each other or produced a lot of artefacts like tiny regions where there were no cells but was marked as a cell. Hence, the main scientific contribution of the project was to make use of mathematical morphology to process the segmented image and produce an image with only regular pixels in the cells. Finally we have the counting step which resulted in the number of cells labeled as 1 or 2. The technical details of all those steps are in the Masters document or in the papers (see references).
Old windows / web version
As the final product of the Masters project, I implemented a windows program in C++ Builder with a very simple interface. Basically the user opens an image and press a button to count. That was the program used by Dr. Alexandre in his laboratory to validate the results. I found the old code and made available in GitHub (see references)
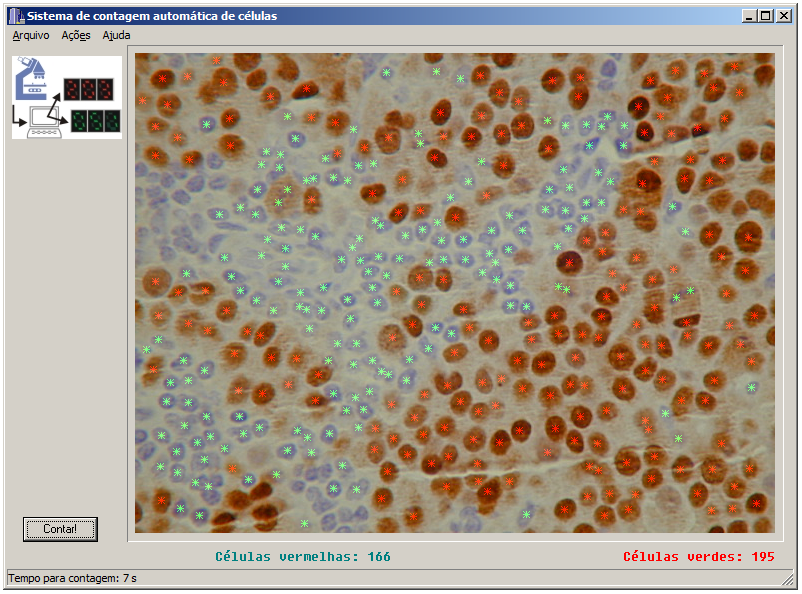
Together with the windows program, I also made an “on-line version” of the application. At the time, an online system that would count cells by uploading an image was a “wonder of technology”😂. Basically it was a web page powered by a PHP script that called a CGI-bin program that did the heavy duty work (implementation of the algorithm) and showed the result. Unfortunately I lost the web page and the CGI code. All I have now is a screenshot that I used in my masters presentation.
iOS
After the Masters defence, for a couple of months I still worked a bit to fix some small stuff and made some small changes here and there. Then, I practically forgot about the project. Dr. Alexandre kept using it for a while but no modifications were made.

Almost 10 years later (around 2012), I was working with iOS development and had the idea of “porting” the program to a mobile platform. Since the iPhone (and iPad) have cameras, maybe we could use to take a picture directly in the objective of the microscope and have the counting right there! So, I did it. I made an app called PathoCounter for iOS that you could take a picture and process the image directly into the device.
The first version was working only on my phone. Just to be a proof of concept (that worked). Then, I realize that the camera on the phone was not the only advance that those 10 years of moore’s law gave me. There was a godzilion of new technology that I could use in the new version of the project! I could use the touch screen to make the interface easy to operate, and allow the user to give more than just the image to the app. The user could paint areas that he knew there were no cells. We could use more than the camera to take images. We could use Dropbox, web, the gallery of the phone, etc. We could produce a report right on the phone and share with other pathologist. The user could have an account to manage his images. And so on… So I did most of what came to my mind and end up with a kind of “high end” cell counting app and made it available in the Apple store. I even put a small animation of the proteins antigens “fitting” in the cell body 🤣. For my surprise, people started to download it!
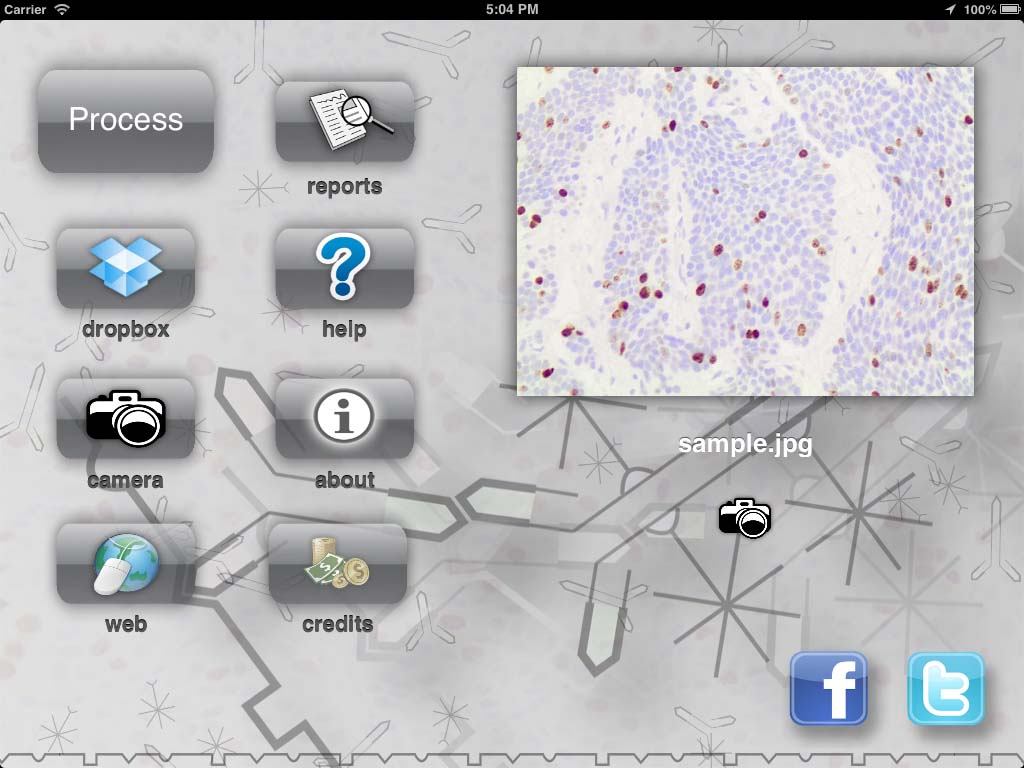
At first I hosted a small web system where the user could login and store images so he could use within the app. I tried to obey some rules for website design but I’m terrible at it. Nevertheless I was willing to “invest my time” to learn how a real software would work. I even learned how to promote a site on facebook and google 😅.
However, I feared that if I had many users the server could not be robust enough, so I tried a crowdfunding campaign at indiegogo.com to buy a good computer to make a private server. For my surprise, some friends baked up the campaign and, in particular, one of my former students Victor Ribas donated $500 dollars! I’m still thankful to him and I was very happy to see such support!! Unfortunately the amount collected wasn’t enough to buy anything different form what I had, so I decided do donate the money to a local Children Hospital that treats children with cancer. Since the goal of the app was to help out with the cancer fight, I figure that the hospital was a good choice for the donation.
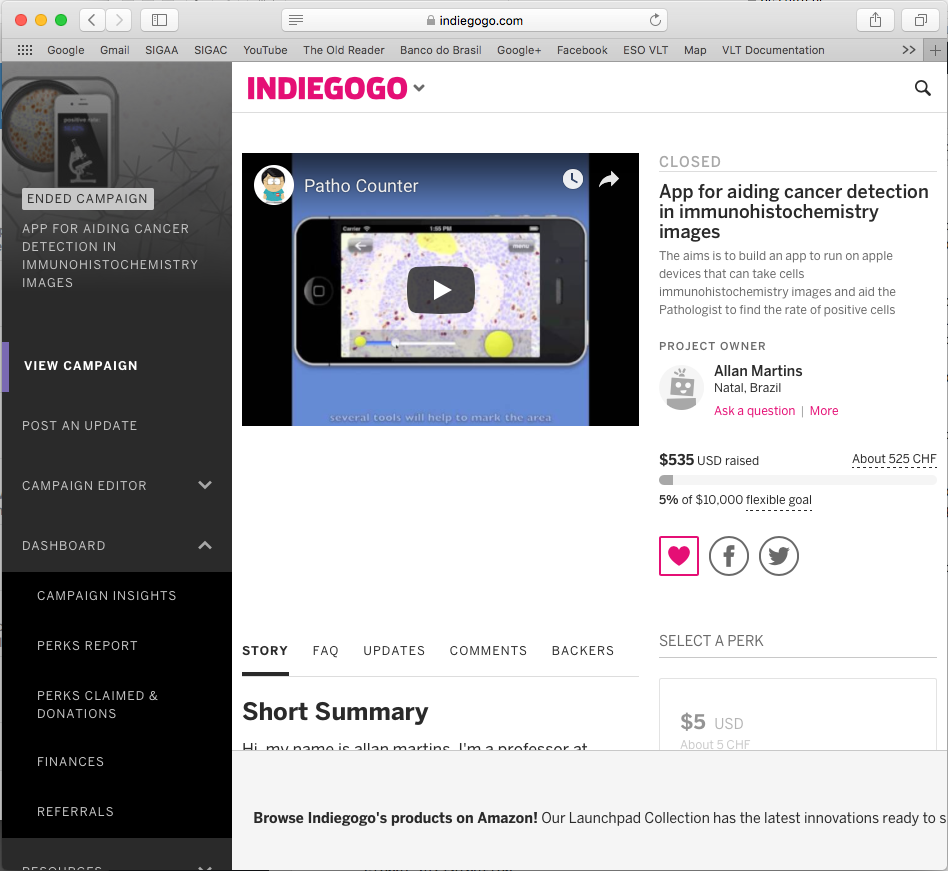
Sadly, this campaign also attracted bad audience. A couple of weeks after the campaign closed and I donated the money, I was accused (formally) of doing unethical fund-gathering for public research. Someone in my department filed a formal accusation against me, and, among some things, told that I was obtaining people’s money from unsuccessful crowdfunding campaigns. That hurts a lot. Even though it was easy to write a defence because I didn’t even kept the money, I had to call the Hospital I did the donation and ask for a “receipt” to attach as a proof in the process. They had offered a receipt at the time I made the donation, but I never imagined that two weeks later I would go though this kind of situation. So, I told them that wasn’t necessary because I was not a legal person or company, and I didn’t want to bother them with the bureaucracy.
Despite this bad pebble in the path, everything that followed was great! I had more the 1000 downloads all around the world. That alone was enough to make me very happy. I also decided to keep my small server running. Some people even got to use the server, although just a handful. Another cool thing that happened was that I was contacted by two pathologists outside Brazil; one from Italy and another from United States. The app was free to download and the user had 10 free exams as a demo. After that the user could buy credits, which nobody did 😂 (I guess because you just could reinstall the app to get the free exams again 😂). I knew about that “bug”, but I was so happy to see that people were actually using the app that I didn’t bother (although Dr. Alexandre told me that I was a “dummy” to not properly monetise the app 🤣).
The app was doing so much more than I expected that I even did some videos explaining the usage. Moreover, I had about 2 or 3 updates. They added some small new features, fixed bugs, etc. In other words, I felt like I was developing something meaningful, and people were using it!
Results
As I said, the main result to me was the usage. To know that people were actually using my app felt great. Actually, the girl from United States that contacted me said that they were using it in dog exams! I didn’t even knew that they did biopsy in animals 😅.
Analytics
At the time I was into the iOS programming “vibe” I use to put a small module in all my apps to gather anonymous analytics data (statistics of usage). One of the data I gathered, when the user permitted of course, was the location were the app was being used. Basically every time you used the software, the app sent the location (without any information or identification of the user whatsoever) and I stored that location. After one or two year of usage, this was the result (see figure).
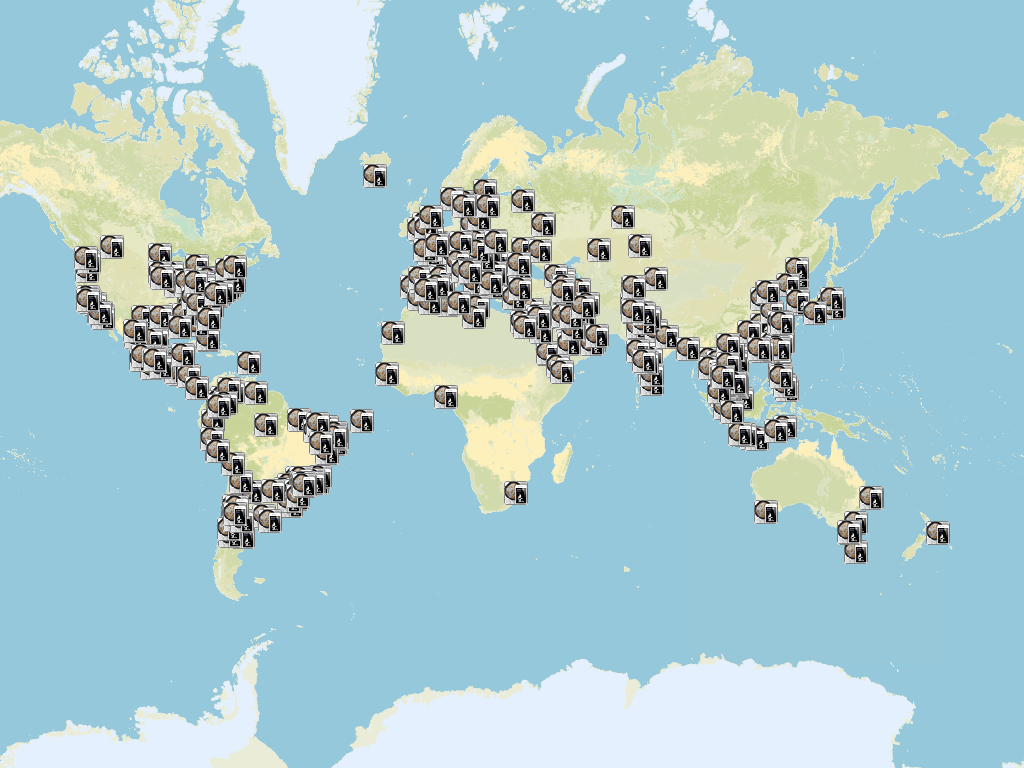
Each icon is a location where the app was used and the user agreed in sharing the location. The image speaks for itself. Basically all the continents were using the app. That is the magic of online stores nowadays. Anybody can have your app being used by, easily, thousands of people! I’m very glad that I lived this experience of software development 😃.
Media
Somehow the app even got the media’s attention. A big local newspaper and one of the public access TV channels here in my city, contacted me. They asked if they could write an article about the app and interview me about it 🤣😂. I told them that I would look bad in front of the camera but I would be very happy to talk about the app. Here goes the first page of the newspaper article in the “Tribuna do Norte” newspaper and the videos of the interviews (the TV Tribuna and TV Universitária UFRN).

Conclusion
I don’t know for sure if this app helped somebody to diagnose an early cancer or helped some doctor to prescribe a more precise dosage of some medicine. However, it shows that everyone has the potential to do something useful. Technology is allowing advances that we could never imagine (even 10 years before when I started my masters). The differences in technology from 2002 to 2012 are astonishing! From a Windows program, going through an online web app, to a worldwide distributed mobile app, software development did certainly changed a lot!
As always, thank you for reading this post. Please, feel free to share it if you like. See you in the next post!
References
Wikipedia – Immunohistochemistry
Sistema de Contagem e Classificação de Células Utilizando Redes Neurais e Morfologia Matemática – Tese de Mestrado
MARTINS, A. M.; DÓRIA NETO, Adrião Duarte ; BRITO JUNIOR, A. M. ; SALES, A. ; JANE, S. Inteligent Algorithm to Couting and Classification of Cells. In: ESEM, 2001, Belfast. Proceedings of Sixth Biennial Conference of the European Sorciety for Engineer and Medicine, 2001.
MARTINS, A. M.; DÓRIA NETO, Adrião Duarte ; BRITO JUNIOR, A. M. ; SALES, A. ; JANE, S. Texture Based Segmentation of Cell Images Using Neural Networks and Mathematical Morphology. In: IJCNN, 2001, Washington. Proceedings of IJCNN2001, 2001.
MARTINS, A. M.; DÓRIA NETO, Adrião Duarte ; BRITO JUNIOR, A. M. ; FILHO, W. A. A new method for multi-texture segmentation using neural networks. In: IJCNN, 2002, Honolulu. Proceedings of IJCNN2002, 2002.
